CSBIO Research
Mosaic
Mechanism Of Action In Compounds
The MOSAIC database is a queryable web interface for browsing chemical genetic interaction data, molecular target predictions based on genetic interactions, Gene Ontology biological process predictions, chemical genetic profile similarity and compound structural similarity. Compounds, genes, and Gene Ontology terms are queryable using MOSAIC. The MOSAIC database is generated from publicly released data from the yeast chemical genomics project
View ProjectneXus
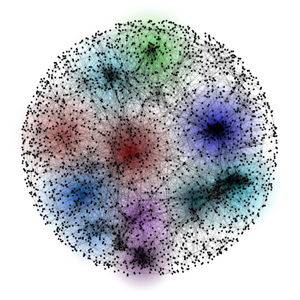
Network - cross(X)-species - Search
neXus is an active subnetwork search algorithm that has been designed to discover conserved subnetworks in the functional linkage networks that are enriched for active genes (such as high foldchange genes in microarray). The algorithm associates statistical significance to each subnetwork that is indicative of how possible such a clustering of genes is when the neXus is run on randomly set of active genes. In the paper [1], the algorithm was used on human and mouse microarray expression data of stem cell with respect to differentiating cells to discover conserved subnetworks representative of stem cell functions in the two species. Variations of the algorithm have been used to discover species specific subnetworks and on single species context. Python implementation of the algorithm is available at http://csbio.cs.umn.edu/neXus/help.html.
View ProjectGRIFn
Gene Relationship Identification in Functional data
GRIFn is a system for evaluation of datasets and methods using a functional genomics gold standard based on curation by expert biolgists. It allows users to assess the ability of their datasets or methods to recapitulate known biology both in a global sense and in the context of specific biological processes. GRIFn allows enables fair comparisons between various data types and methods.
View ProjectbioPIXIE
Biological Pathway Inference from eXperimental Interaction Evidence
bioPIXIE is a novel system for biological data integration and visualization for S. cerevisiae. It allows the user to discover interaction networks and pathways in which the user's gene(s) of interest participate. The system is based on a Bayesian algorithm for identification of biological networks based on integrated diverse genomic data.
View ProjectChARMview
Chromosomal Abberation Region Miner and Viewer
ChARMView is a visualization and analysis system for guided discovery of chromosomal abnormalities from microarray data. Our system facilitates manual or automated discovery of aneuploidies through dynamic visualization and integrated statistical analysis. ChARMView can be used with array CGH and gene expression microarray data, and multiple experiments can be viewed and analyzed simultaneously.
View ProjectGOLEM
Gene Ontology Local Exploration Map
GOLEM is a tool for viewing, navigating, and analyzing the hierarchical structure and annotations to the gene ontology. The visualization component allows a user to see the local graph structure around a GO term of interest and navigate to nearby nodes. GOLEM also provides the ability to look for statistical enrichment of GO terms in lists of genes and then observe the relationships between those terms. GOLEM is available both as an applet for use online and as a standalone download.
View Project